Andreas Nienkötter received a master’s degree in computer science from University of Münster, Germany, in 2015, followed by a Ph.D. in 2021. He is currently in a Postdoctoral position in Prof. X. Jiang’s Research Group for Pattern Recognition and Image Analysis. His research interests include consensus learning using the generalized median, vector space embedding methods, and dimensionality reduction methods.
ULTRACEPT researcher Andreas Nienkötter from the University of Münster, Germany recently published a journal article titled ‘Kernel-based generalized median computation for consensus learning’ in IEEE Transactions on Pattern Analysis and Machine Intelligence, 2022, doi: 10.1109/TPAMI.2022.3202565. The journal covers research in computer vision and image understanding, pattern analysis and recognition, machine intelligence, machine learning, search techniques, document and handwriting analysis, medical image analysis, video and image sequence analysis, content-based retrieval of image and video, and face and gesture recognition.
Research Summary
Consensus learning is an important field in computer science with application in many diverse fields. The task is to compute a single representative of a given set of singular data or results, e.g. a final prediction from a set of input predictions, an averaged rotation or position from several measured rotations or positions, or a representative DNA-sequence computed from several faulty or mutated DNA sequences.
The generalized median is a very intuitive method to compute such a consensus related to the standard median of numbers. The median of numbers is not only the value in the centre of the sorted list, but also a value that minimizes the sum of absolute differences to the values in the list.
Transferred to general domains such as positional data, genetic strings, rotation matrices, physical signals, the generalized median is an object which minimizes the sum of distance to the input set given a domain specific distance. Surprisingly, the computation of the generalized median is often NP-hard or even worse, even for simple objects such as two-dimensional vectors with the euclidean distance. Nevertheless, the generalized median is popular in practice as it is much more robust against outliers compared to simple averaging.
In this research, we developed a method to compute the generalized median in any domain requiring only minimal user input by computing the median in kernel space. The system is based on the distance-preserving embedding method used in our previous work, as seen in Figure 1.
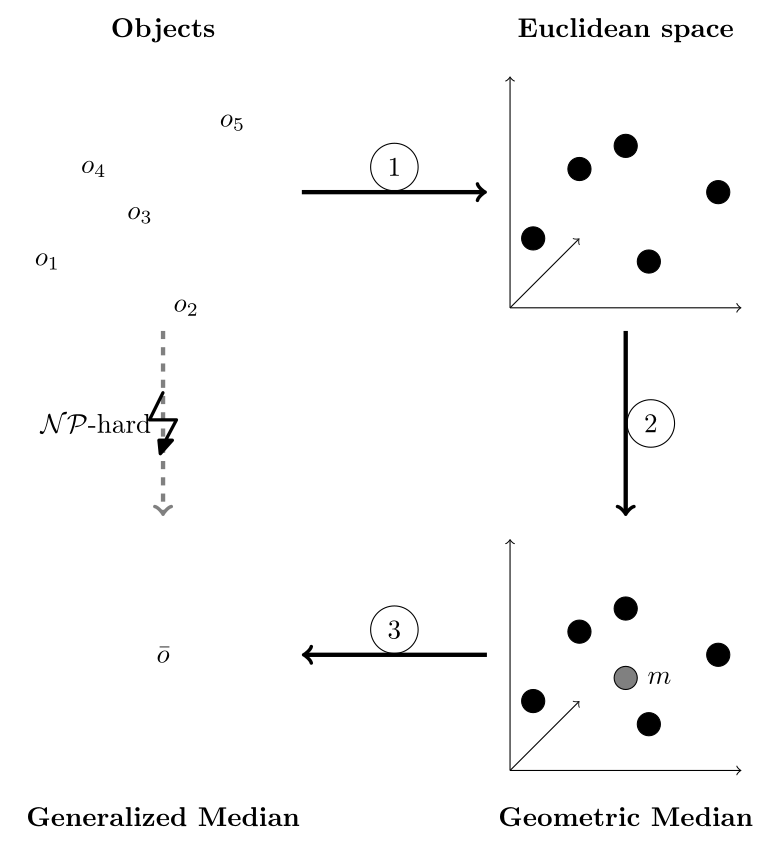
The distance preserving embedding framework computes the generalized median in three steps. 1) The objects are embedded into a high-dimensional vector space so that the pairwise distances between vectors are approximately the same as the pairwise distances between the input objects. 2) The generalized median of these vectors (often called geometric median) is approximated in vector space, for which fast algorithms exist. 3) Finally, the median is reconstructed by combining the objects corresponding to the neighbours of the median in vector space.
Research Highlights
While this previous framework has been shown to accurately compute a generalized median, it has a major drawback: the embedding into an euclidean vector space. Unfortunately, most distances (e.g. the string edit distance) cannot be accurately represented in an euclidean vector space, leading to distortions in the position of vectors and ultimately to a less accurate median computation. To overcome this, the kernel-based median computation in this work removes the need for an explicit vector space entirely by using kernel functions instead.
As we were able to show in our work, the above framework does not actually need the explicit vectors of objects or even the explicit vector position of the generalized median. The scalar product between these vectors suffices instead. These scalar products can be efficiently computed using kernel functions. Kernel functions are defined as functions computing the scalar product of objects after transformation into a Hilbert space. Notably, this space can be unknown and even infinite-dimensional, something that was not possible before with explicit embedding methods. Some of these kernel functions only require the distances between objects, meaning that there is no additional input required by the user. These easy to use kernel functions even compute the scalar product in a pseudo-euclidean space leading to a more accurate representation of distances, something that was not possible with an euclidean vector space before.
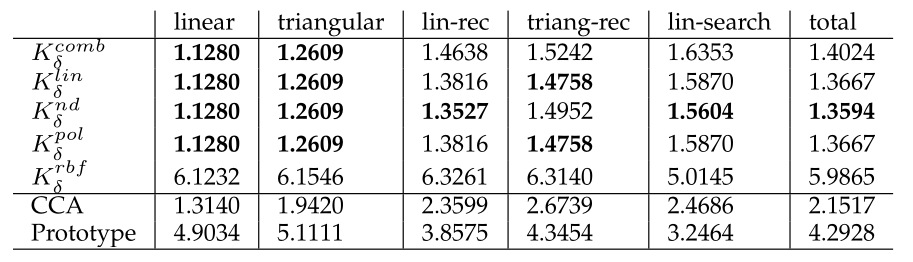
As seen in Figure 2, we were able to achieve superior results using this method compared to previous methods in string, clustering and ranking data. The results were computed with less parameters in a similar time.
Generalized Median Toolbox
The kernel-based generalized median method as well as the previous distance preserving embedding methods are available to the public as python toolbox (https://pypi.org/project/gmtb/). We invite anyone interested in this topic to try it in their consensus computation tasks.
